T7-ORACLE: The Protein Engineering Tool Set to Transform Drug Discovery
Discover T7-ORACLE, the revolutionary protein engineering platform accelerating evolution by 100,000x. Learn how this innovation, merging continuous evolution with AI, is reshaping the future of biotech and medicine.

Written by Lavanya, Intern, Allegedly The News
LA JOLLA, California, August 11, 2025
In a move poised to fundamentally reshape the landscape of drug discovery and biotechnology, scientists at Scripps Research have unveiled a groundbreaking protein engineering platform named T7-ORACLE. Described in a recent publication in the journal Science, this toolset promises to accelerate the process of protein evolution by a staggering factor of 100,000, compressing what would typically take years of laborious lab work into a matter of days or weeks. This is not just an incremental improvement; it is a paradigm shift that could unlock solutions to some of humanity's most pressing challenges, from curing intractable diseases to developing eco-friendly industrial processes.
The Unprecedented Leap: What is T7-ORACLE?
For decades, scientists have relied on directed evolution to engineer proteins with new or enhanced functions. This process, which mimics natural selection in a lab setting, involves generating a library of protein variants through mutagenesis and then screening them to find the ones with desired properties. However, this method is notoriously slow and resource-intensive, with each "round" of evolution often taking weeks. The sheer scale of the protein sequence space, the astronomical number of possible amino acid combinations, has been a formidable barrier, making it impossible to explore more than a minuscule fraction of the possibilities.
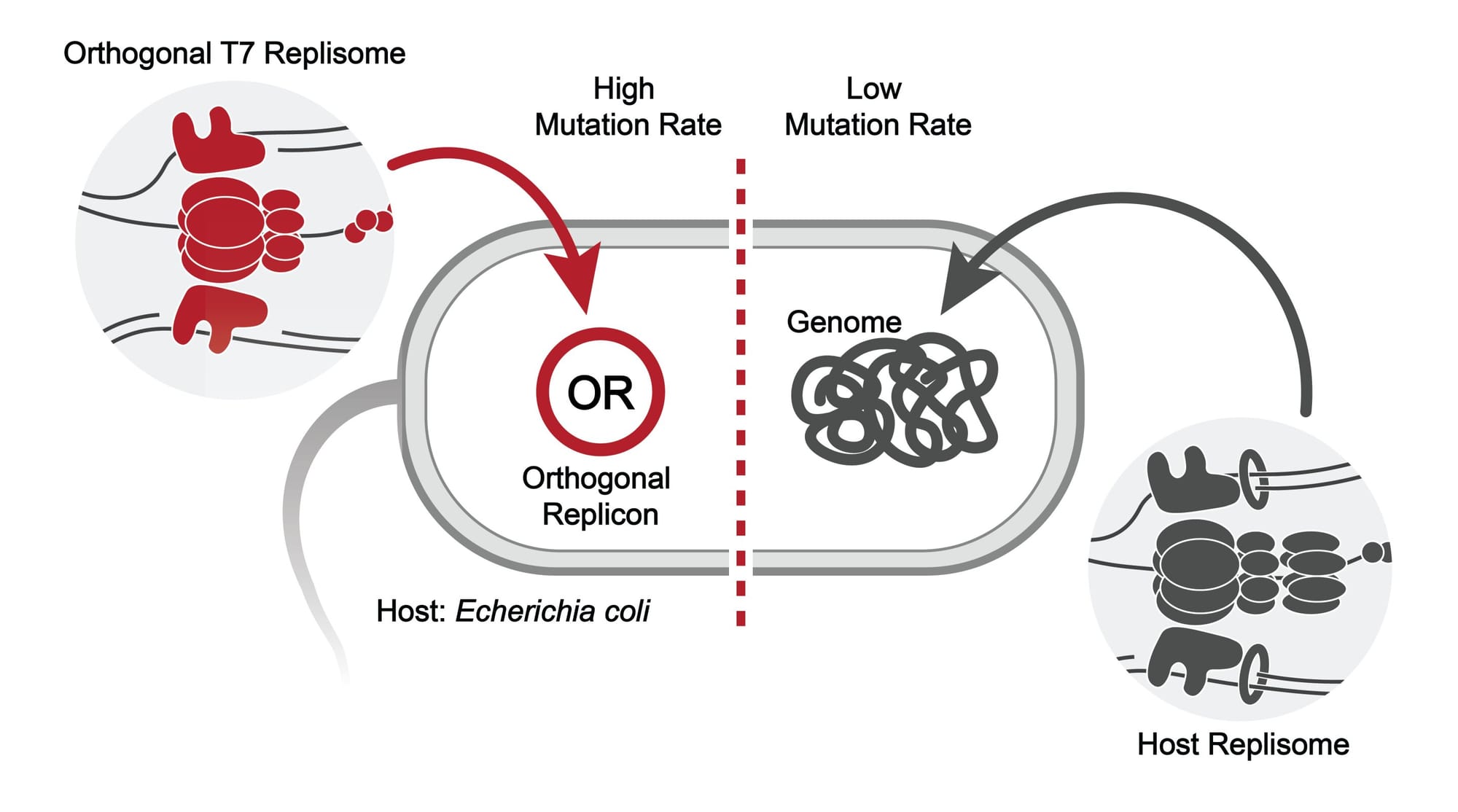
T7-ORACLE, a name combining the T7 bacteriophage with the concept of a predictive "oracle," shatters this barrier. At its core, the technology is a self-contained, orthogonal replication system operating within standard E. coli bacteria. This means it has its own, separate DNA replication machinery that functions independently of the cell's main genome. The key to its speed is a modified T7 DNA polymerase, an enzyme engineered by the Scripps team to be deliberately error-prone. This "hypermutation" causes the target gene, carried on a small circular piece of DNA called a plasmid, to mutate at a rate 100,000 times higher than the natural background mutation rate.
Critically, this high mutation rate is confined to the plasmid, leaving the host bacterium's own genome untouched and healthy. As the bacteria grow and divide, the target protein gene is continuously evolving, with each cell division representing a new round of evolution. Since E. coli can divide every 20 minutes, this translates to a rapid-fire, continuous evolutionary process that runs with minimal human intervention.
A Demonstration of Power: Evolving Antibiotic Resistance
To prove the platform's capabilities, the Scripps team undertook a powerful proof-of-concept experiment. They inserted a common antibiotic resistance gene, TEM-1 β-lactamase, into the T7-ORACLE system. They then exposed the E. coli cells to progressively higher concentrations of various antibiotics. The results were astounding. In less than a week, the system evolved versions of the enzyme that could resist antibiotic levels up to 5,000 times higher than the original.
What was particularly remarkable was that the mutations observed in the lab closely mirrored those found in clinical settings where bacteria have naturally evolved resistance over many years. This real-world relevance not only validated T7-ORACLE's power but also demonstrated its potential as a tool for understanding and predicting the evolution of drug resistance in pathogens. It also hinted at the possibility of designing new drugs that can overcome existing resistance mechanisms before they even emerge.
T7-ORACLE's Transformative Role in Drug Discovery
The pharmaceutical industry, with its long and expensive drug development pipelines, is perhaps the biggest beneficiary of this breakthrough. The ability to rapidly engineer and optimize proteins could collapse the timelines for developing new therapeutics.
- Rapid Development of Biologics: T7-ORACLE can be used to evolve highly specific and potent antibodies, therapeutic enzymes, and other protein-based drugs. For example, it could accelerate the development of broadly neutralizing antibodies against rapidly mutating viruses like influenza or coronaviruses, creating a new paradigm for pandemic preparedness.
- Engineering Cancer Therapies: Researchers are already using the platform to evolve proteases, enzymes that break down other proteins, to specifically target and degrade proteins associated with cancer and neurodegenerative diseases. This could lead to a new generation of highly targeted therapies with fewer side effects.
- Precision Medicine: The platform's adaptability means it could be used to engineer therapeutic proteins tailored to an individual patient’s unique genetic profile or disease subtype, a key goal of precision medicine.
- Overcoming Drug Resistance: As demonstrated in the proof-of-concept study, T7-ORACLE can be a powerful tool for proactively designing drugs that are less susceptible to resistance.
The traditional drug development process involves a long and arduous journey of lead discovery, optimization, and clinical trials. T7-ORACLE has the potential to dramatically shorten the initial discovery and optimization phases, allowing scientists to generate and test hundreds or thousands of improved candidates in a fraction of the time. This reduction in the discovery timeline could translate into significant cost savings and, more importantly, faster access to life-saving medicines for patients.
From Pharmaceuticals to Planetary Health: Cross-Industry Applications
While the impact on medicine is immediate and profound, the utility of T7-ORACLE extends far beyond the clinic. Enzymes are the workhorses of nature and industry, and the ability to rapidly engineer them opens up a world of possibilities.
- Sustainable Industrial Processes: Industrial biotechnology relies heavily on enzymes to produce everything from biofuels and bioplastics to detergents and food additives. T7-ORACLE can be used to evolve enzymes that are more stable, active, and efficient under industrial conditions, such as high temperatures or in the presence of harsh chemicals. This leads to more cost-effective and environmentally friendly manufacturing.
- Environmental Remediation: Engineered enzymes can be designed to break down persistent environmental pollutants, such as plastics, pesticides, or oil. T7-ORACLE could accelerate the development of "super-enzymes" for bioremediation, enabling faster and more effective cleanup of contaminated sites.
- Agriculture and Food Production: The toolset could be used to create enzymes that improve crop yields, enhance nutritional content, or act as natural biopesticides. It could also lead to the development of new enzymes for food processing that improve taste, texture, and shelf life.
A Synergistic Future: T7-ORACLE and the AI Revolution
The true power of T7-ORACLE is not just in its speed, but in its synergy with the rapidly advancing field of artificial intelligence. AI is already being used to predict protein structures and design novel protein sequences. However, these predictions often require extensive experimental validation. T7-ORACLE provides the perfect "testbed" for these AI-driven hypotheses.
- AI-Guided Evolution: Instead of purely random mutations, AI models can suggest specific amino acid changes most likely to improve a protein's function. T7-ORACLE can then be used to rapidly test these targeted mutations, generating a flood of high-quality data that can be fed back into the AI model for further refinement. This creates a powerful, self-improving loop of computational design and experimental validation.
- Data Analysis on an Unprecedented Scale: The vast amount of data generated by T7-ORACLE's ultra-high-throughput screening would be impossible for humans to analyze. AI and machine learning algorithms can sift through this data, identifying patterns and insights that guide the evolutionary process more intelligently and efficiently.
This dynamic interplay between AI and ultra-fast evolution represents a new frontier in biotech, where rational design and random mutation are no longer mutually exclusive but are combined to achieve results far beyond what either could accomplish alone.
The Innovation Journey: From Idea to Impact
The development of T7-ORACLE was the culmination of years of dedicated work by a team led by Professor Christian Diercks at Scripps Research. The journey was a classic example of synthetic biology—the design and construction of new biological parts, devices, and systems.
The innovation was built on the foundation of earlier "orthogonal replication" systems that operated independently of a cell's main machinery. However, these systems had limitations in their mutation rates, growth speed, or ease of use. The Scripps team’s breakthrough was in successfully engineering the T7 bacteriophage's replication system to be both highly error-prone and seamlessly integrated into the fast-growing E. coli platform. This technical elegance, combined with the modularity of the system, is what makes T7-ORACLE a game-changer. The platform is designed to be user-friendly, compatible with standard lab workflows, and customizable to evolve virtually any protein of interest.
The Big Picture: A Turning Point in Biotech History
The excitement surrounding T7-ORACLE is understandable. It addresses a fundamental bottleneck in biological research and engineering. Before this, the pace of protein evolution was a rate-limiting step in countless scientific endeavors. By accelerating this process by five orders of magnitude, the tool enables researchers to ask entirely new kinds of questions and pursue projects that were previously considered impossible due to time constraints.
It represents a major step towards making the power of evolution a predictable, programmable, and accessible tool for human innovation. Experts see this as a pivotal moment, much like the advent of CRISPR-Cas9 for gene editing, that will catalyze a new wave of discoveries and applications across all life sciences.
From Tinkering to True Engineering
For decades, protein engineering has been a blend of rational design and laborious trial-and-error. With T7-ORACLE, we are moving from a world of tinkering to one of true engineering. This revolutionary platform transforms protein evolution from a slow, serendipitous process into a fast, iterative, and programmable one. By providing an "evolution engine" that can be directed and controlled, it empowers scientists to design and build the molecular tools needed to solve the great challenges of our time. The future of medicine, industry, and environmental sustainability will, in no small part, be shaped by the proteins we can now so rapidly and elegantly create.
Pondering the Future
The emergence of such a powerful technology invites important discussion. As we stand on the precipice of this new era, we must consider its broader implications and the questions it raises for science and society. As a tool that can be used with standard lab workflows, T7-ORACLE has the potential to democratize protein engineering. How can we ensure that this technology is accessible to a wide range of researchers and institutions, preventing its power from being concentrated in a few hands?
Sources
ScienceDaily, Scripps Research official press releases, Technology Networks, Shutterstock, and other verified scientific publications.




